2018
Bouhaddou, 2018, PLoS Comp Bio : A mechanistic model to interpret context-specific landscapes between driver pathways and cell fates, providing a framework for designing more rational cancer combination therapy. Download the model here
2017
Barrette, 2017, ACS Chem. Neuroscience : A mechanistic ODE model of pan-cancer driver pathways that simulates how varied doses of microenvironmental signals and brain-penetrant kinase inhibitor combinations influence the dynamics of stochastic proliferation and death in single glioma cells. The model is tailored to 14 different glioma patients from The Cancer Genome Atlas to form a "virtual patient cohort".
Download the model here
2015
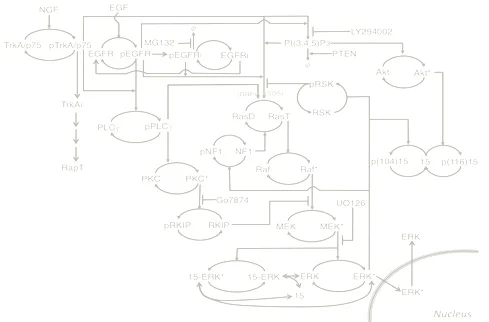
Birtwistle, 2015, Interface : An ODE model that considers combinatorial complexity of phosphorylation, dephosphorylation and protein recruitment events in EGF signaling, given in (i) BioNetGen language and (ii) a model reduction method in MATLAB.
Download the model here
2014
Zhang et al., 2014, CPT-PSP : A mechanistic ODE model of VEGF signaling to ERK and Akt, which includes pharmacokinetic models of many drugs for targets in this pathway. The model is wrapped around an optimization framework for proposing dosing and treatment frequency strategies.
Download the model here
Bouhaddou et al., 2014, Mol Biosystems : A comprehensive mechanistic ODE model of receptor ligand binding and dimerization mechanisms, with extensive parametric variation.
Download the model here
2013
Birtwistle et al., 2013, CPT-PSP : Instantiations of the 2007 ErbB model (above) used to demonstrate our viewpoint that mechanistic models have particularly useful features in looking forwards to precision medicine applications.
Download the model here
2012
Birtwistle et al., 2012, Mol Biosystems : Code that evaluates the overlap probability of protein levels two populations with different means; e.g. the probability that a cell a si/shRNA population might have by chance higher levels than a cell in the control population, despite adequate mean knockdown.
Download the model here
Birtwistle et al., 2012, BMC Sys Biol : A mechanistic ODE model of signal transduction EGF-activated Ras to ERK, which includes descriptions of stochastic gene expression as variable initial conditions.
Download the model here
2010
Nakakuki et al., 2010, Cell : Three models are included. Two are mechanistic ODE models that describe signal transduction EGF or HRG-stimulated MEK activity to c-Fos production and activation. One is a simplified semi-empirical version of the same, which we call the core model.
Download the model here
Sturm et al., 2010, Sci Signal : An ODE model developed by Richard Orton to describe negative feedback in the ERK/MAPK pathway.
Download the model here
2009
von Kreigshiem et al., 2009, Nat Cell Biol : An ODE model of EGF and NGF signaling to ERK and Akt in PC-12 cells including new signaling mechanisms identified by interaction proteomics.
Download the model here
2007
Birtwistle et al., 2007, Mol Syst Biol : A mechanistic ODE model of the ErbB receptor signaling network EGF and HRG to ERK and Akt in MCF-7 cells.
Download the model here